Differential Chromatin Interaction
Do the DCI(differential chromatin interaction) analysis between two Hi-C matrics. The Selfish method used here is described in the paper:
Abbas Roayaei Ardakany, Ferhat Ay, Stefano Lonardi, Selfish: discovery of differential chromatin interactions via a self-similarity measure, Bioinformatics, Volume 35, Issue 14, July 2019, Pages i145–i153, https://doi.org/10.1093/bioinformatics/btz362

[1]:
import coolbox
from coolbox.api import *
[2]:
coolbox.__version__
[2]:
'0.4.0'
[3]:
data_dir = "../../../tests/test_data/"
cl1 = f"{data_dir}/cool_chr1_89000000_90400000_for_cmp_1.mcool"
cl2 = f"{data_dir}/cool_chr1_89000000_90400000_for_cmp_2.mcool"
region = "chr1:89237500-90227500"
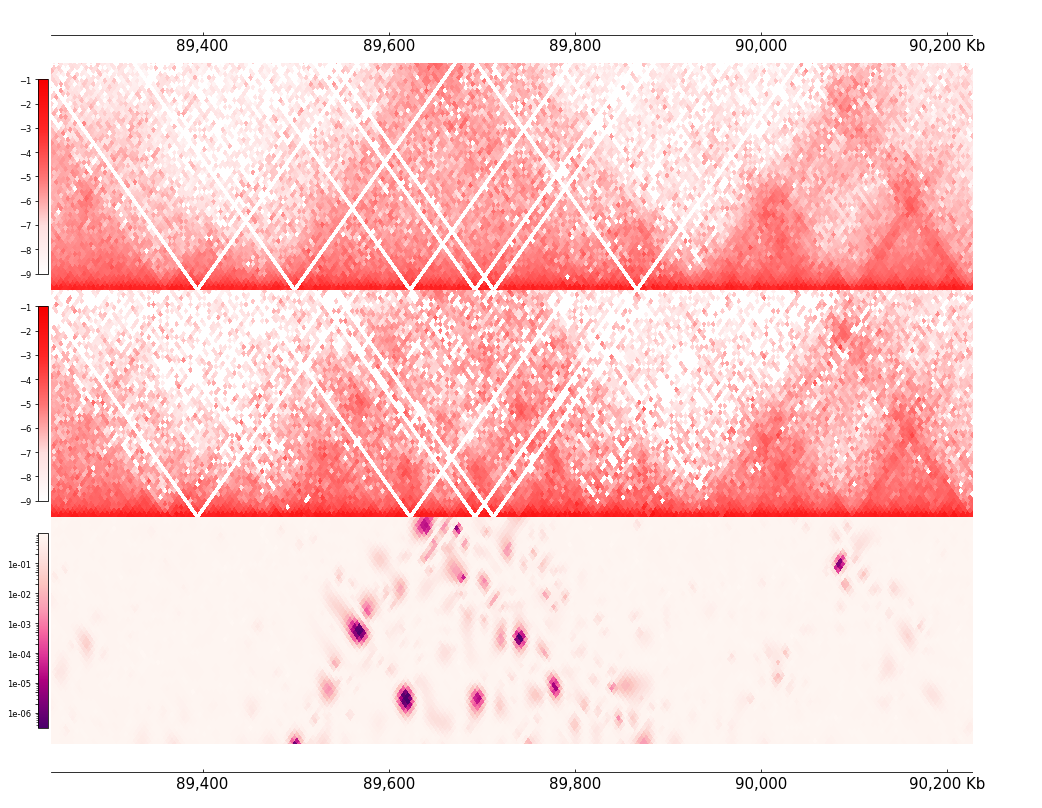
The Selfish track input two hic track(Cool or DotHiC), it perform the selfish algorithm on the local fetched matrics. Output the q-value(p-value after fdr_bh correction) of differential interaction analysis:
[4]:
with MinValue(-9), MaxValue(-1):
cool1 = Cool(cl1)
cool2 = Cool(cl2)
frame = XAxis() + \
cool1 + \
cool2 + \
Selfish(cool1, cool2, norm="log") + \
XAxis()
frame *= Feature(depth_ratio=0.35)
frame.plot(region)
[4]:
CLI code
[5]:
%%bash
data_dir="../../../tests/test_data/"
cl1="${data_dir}/cool_chr1_89000000_90400000_for_cmp_1.mcool"
cl2="${data_dir}/cool_chr1_89000000_90400000_for_cmp_2.mcool"
coolbox add XAxis - \
start_with Feature --depth_ratio="0.35" - \
start_with Feature --min_value="-9" --max_value="-1" - \
add Cool $cl1 - \
add Cool $cl2 - \
end_with - \
add Selfish $cl1 $cl2 --norm 'log' - \
end_with - \
add XAxis - \
goto "chr1:89237500-90227500" - \
plot /tmp/test_coolbox_selfish.png