TADs
Plot TAD(topologically associated domains)s in upon Hi-C contact matrix.
[1]:
import coolbox
from coolbox.api import *
[2]:
coolbox.__version__
[2]:
'0.4.0'
TAD file should store as BED6 format:
[3]:
!head -n 10 ../../../tests/test_data/tad_chr9_4000000_6000000.bed
[4]:
example_cool = "../../../tests/test_data/cool_chr9_4000000_6000000.mcool"
example_tads = "../../../tests/test_data/tad_chr9_4000000_6000000.bed"
test_region = "chr9:4000000-6000000"
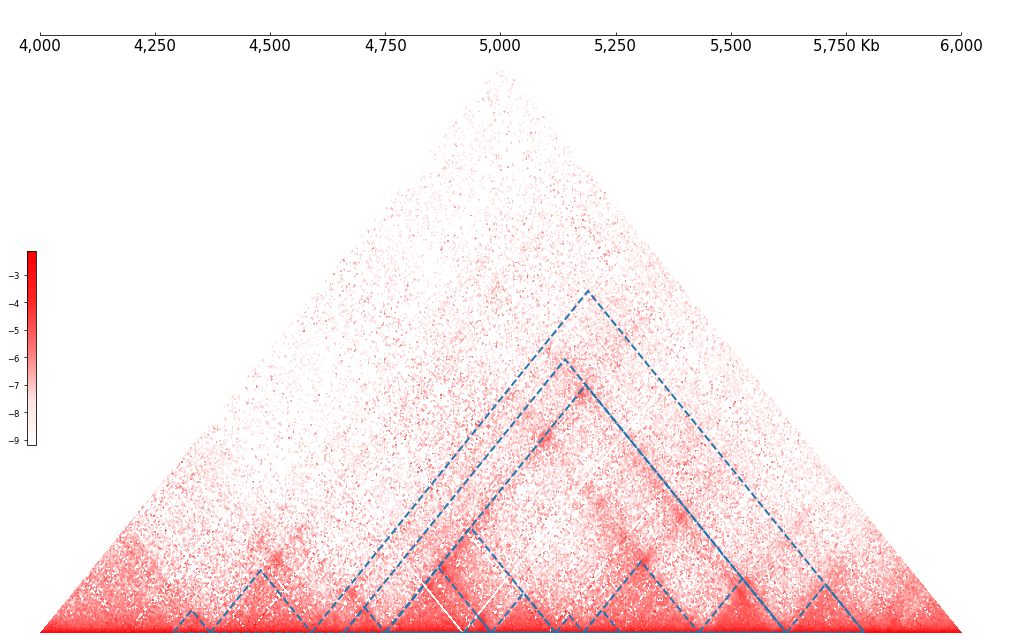
frame = XAxis() + \
Cool(example_cool) + \
TADCoverage(example_tads, border_only=True, alpha=1.0)
frame.plot(test_region)
[4]:
[5]:
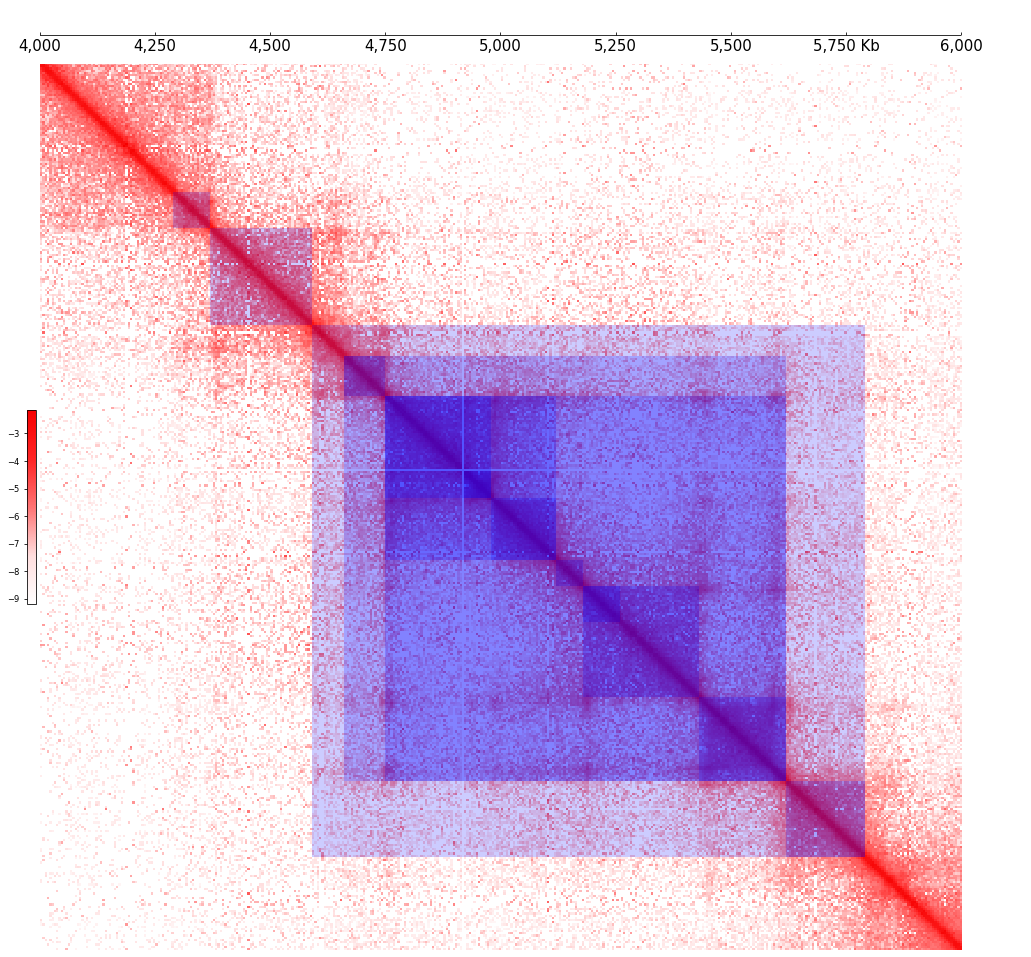
frame = XAxis() + \
Cool(example_cool, style="matrix") + \
TADCoverage(example_tads, alpha=0.2, color="blue")
frame.plot(test_region)
[5]:
CLI code
[6]:
%%bash
cool="../../../tests/test_data/cool_chr9_4000000_6000000.mcool"
tads="../../../tests/test_data/tad_chr9_4000000_6000000.bed"
coolbox add XAxis - \
add Cool $cool - \
add TADCoverage $tads --border_only True --alpha 1.0 - \
goto "chr9:4000000-6000000" - \
plot /tmp/test_coolbox.png
[7]:
%%bash
cool="../../../tests/test_data/cool_chr9_4000000_6000000.mcool"
tads="../../../tests/test_data/tad_chr9_4000000_6000000.bed"
coolbox add XAxis - \
add Cool $cool --style matrix - \
add TADCoverage $tads --color blue --alpha 0.2 - \
goto "chr9:4000000-6000000" - \
plot /tmp/test_coolbox.png